
Similarly, the transferred segments need to exhibit the donor's signature and to be significantly different from the recipient's.
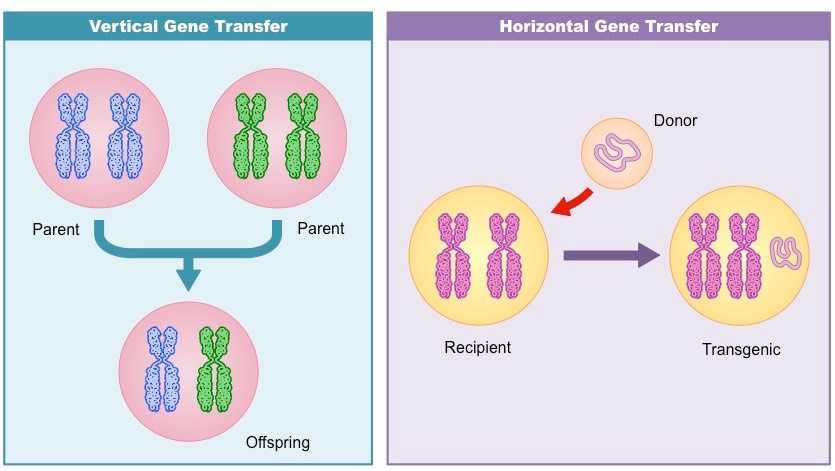
However, because they rely on the uniformity of the host's signature to infer HGT events, not accounting for the host's intragenomic variability will result in overpredictions-flagging native segments as possible HGT events. It has been a considerable advantage at the early times of the sequencing era, when few closely related genomes were available for comparative methods. The main feature of parametric methods is that they only rely on the genome under study to infer HGT events that may have occurred on its lineage. Implicit phylogenetic methods bypass gene tree reconstruction, e.g., by looking at discrepancies between pairwise distances between genes and their corresponding species. Explicit phylogenetic methods reconstruct gene trees and infer the HGT events likely to have resulted into that particular gene tree. (2) Phylogenetic approaches rely on the differences between genes and species tree evolution that result from HGT. Regions with atypical values are inferred as having been horizontally transferred. (1) Parametric methods infer HGT by computing a statistic, here GC content, for a sliding window and comparing it to the typical range over the entire genome, here indicated between the two red horizontal lines. Conceptual overview of HGT inference methods. Phylogenetic methods can be further divided into those that reconstruct and compare phylogenetic trees explicitly and those that use surrogate measures in place of the phylogenetic trees. Phylogenetic methods examine evolutionary histories of genes involved and identify conflicting phylogenies. Parametric methods search for sections of a genome that significantly differ from the genomic average, such as guanine-cytosine (GC) content or codon usage. These methods can be broadly separated into two groups: parametric and phylogenetic methods ( Fig 1). To infer HGT events, which may not necessarily result in phenotypic changes, most contemporary methods are based on analyses of genomic sequence data. Similar observations in the 1940s and 1950s showed evidence that conjugation and transduction are additional mechanisms of horizontal gene transfer. Showing that virulence was able to pass from virulent to nonvirulent strains of Streptococcus pneumoniae, Griffith demonstrated that genetic information can be horizontally transferred between bacteria via a mechanism known as transformation. In view of the use of GM crops and microbes in agricultural settings, in this mini-review we focus particularly on the presence and role of MGE in soil and plant-associated bacteria and the factors affecting gene transfer.Horizontal gene transfer (HGT) was first observed in 1928, in Frederick Griffith’s experiment. This review summarizes the present state of knowledge on HGT between bacteria as a crucial mechanism contributing to bacterial adaptability and diversity. Thus, for a risk assessment it is important to understand the extent of HGT and genome plasticity of bacteria in the environment. Second, the genetically modified bacterium released into the environment might capture mobile genetic elements (MGE) from indigenous micro-organisms which could extend its ecological potential. First, introduced genetic sequences from a genetically modified bacterium could be transferred to indigenous micro-organisms and alter their genome and subsequently their ecological niche. The occurrence of HGT among bacteria in the environment is assumed to have implications in the risk assessment of genetically modified bacteria which are released into the environment. Horizontal gene transfer (HGT) refers to the acquisition of foreign genes by organisms.


 0 kommentar(er)
0 kommentar(er)
